Myeloma Genomics and Microenvironment and immune profiling
Category: Myeloma Genomics and Microenvironment and immune profiling
Characterizing the Molecular Impact of DIS3 Mutation-Driven Mechanisms in Multiple Myeloma
(PA-227) Characterizing the Molecular Impact of DIS3 Mutation-Driven Mechanisms in Multiple Myeloma

Ceanne Elliott, B.S.
Ph.D. Candidate
Indiana University School of Medicine
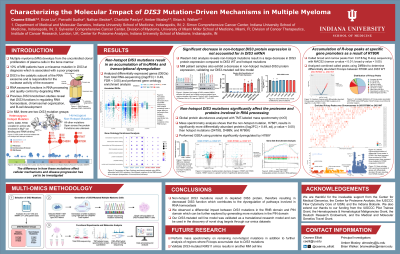
DIS3 is the main catalytic subunit of the RNA exosome—a critical complex responsible for degrading most cellular RNA. Mutations in DIS3 occur in 10% of multiple myeloma (MM) patients and can be either recurrent heterozygous hotspot mutations or homozygous non-hotspot mutations, both associated with poor prognosis. Determining the distinct mechanisms by which these mutation subtypes impair RNA exosome function and drive MM pathogenesis is critical for unraveling their biological role in disease progression and advancing targeted therapeutic strategies.
Methods:
We analyzed patient samples from the Multiple Myeloma Research Foundation (MMRF) CoMMpass Study (n=1,245; DNA and RNA sequencing), combined with Indiana University samples (SMM, n=14; NDMM, n=36; RRMM, n=54; DNA and RNA sequencing), to determine DIS3 mutation rates and downstream effects. Endogenous DIS3 was mutated in both KMS11 and JJN3 cell lines using CRISPR-Cas9 homology directed repair, and mutated colonies were isolated for 4 non-hotspot mutations (Y106C, T262P, L544P, H788Y), 3 hotspot mutations (D479G, D488N, R780K), and controls. We examined the functional impact of these DIS3 mutations via Western blot analysis, total RNA-sequencing for differentially expressed gene (DEG) and pathway analysis, R-loop detection, tandem mass spectrometry (LC-MS/MS), and Olink Explore 3072.
Results:
DEG analysis of MMRF CoMMpass RNA-sequencing data between subsampled DIS3 hotspot (n=13) and non-hotspot (n=24) groups with controls (n=24) resulted in a substantial increase (p< 0.0001) of lncRNAs—where non-hotspot mutations had a significantly larger increase than hotspot mutations. This result was also observed in our DIS3-mutated cell lines, therefore validating our experimental approach. Over half of the DEGs in mutated cell lines were found to be regulated by AU-rich elements, suggesting aberrant post-transcriptional regulation contributing to disease pathology. The introduction of endogenous DIS3 mutations did not affect DIS3 mRNA expression, but there was a 25-fold decrease (p< 0.0001) in DIS3 protein expression only in non-hotspot mutated cells, validated by Western blot analysis of MM patient CD138+ samples. In contrast, hotspot mutations exhibited no DIS3 protein abundance change in both our DIS3-mutated cell lines and patient samples, as confirmed by mass spectrometry. Lastly, LC-MS/MS analysis revealed non-hotspot mutations resulted in the significant (p< 0.05) dysregulation of histone linkers (H1.2-H1.5, H1-10) which may be contributing to the overall downregulation of proteins and transcriptional pathways seen in proteomic and pathway analysis.
Conclusions:
Our findings show DIS3 mutations in MM have unique molecular consequences by subtype, affecting both RNA processing and exosome function. These results advance our understanding of how DIS3 mutations contribute to disease progression and emphasize the importance of considering mutation-specific effects when developing targeted therapies.
