Myeloma Genomics and Microenvironment and immune profiling
Category: Myeloma Genomics and Microenvironment and immune profiling
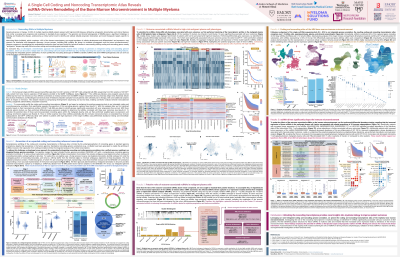
A Single-Cell Coding and Noncoding Transcriptomic Atlas Reveals ncRNA-Driven Remodeling of the Bone Marrow Microenvironment in Multiple Myeloma
(PA-251) A Single-Cell Coding and Noncoding Transcriptomic Atlas Reveals ncRNA-Driven Remodeling of the Bone Marrow Microenvironment in Multiple Myeloma

Marina E. Michaud, MSc (she/her/hers)
PhD Candidate, Cancer Biology
Emory University School of Medicine
Interactions between myeloma cells and the immune microenvironment are a critical target for developing next-generation immunotherapies for multiple myeloma (MM). To investigate these interactions, single-cell transcriptomics enables comprehensive profiling of the MM bone marrow microenvironment (BMME); however, current analyses primarily focus on coding genes, neglecting the substantial noncoding transcriptome due to challenges in genomic overlap. To address this, we have developed a novel integrated coding-noncoding RNA profiling approach to elucidate the noncoding landscape of the MM-BMME. We hypothesize that specific cytogenetic mutations give rise to distinct genomic profiles regulated by the noncoding transcriptome, thereby altering MM phenotypes and, consequently, the BMME.
Methods:
To accomplish this, we generated an expanded human reference genome integrating mRNA and ncRNA annotations ( >175k genes, 80% noncoding) from LncBook2.1 and GENCODEv47, maximizing noncoding gene capture while preserving coding gene detection from single-cell or bulk sequencing data. Benchmarking this approach, we demonstrate that alignment with the integrated genome annotation increases noncoding gene capture by over threefold (to 133,846 genes) in a single-cell dataset.
Harnessing our integrated annotation, we generated and aligned single-cell RNA-seq data (N=481) from CD138- immune cells, as well as bulk RNA-seq data from CD138+ myeloma cells, from patients enrolled in the MMRF CoMMpass trial. This resulted in the first integrated coding-noncoding single-cell atlas of MM-BMME comprising >1.9 million cells, spanning plasma, stromal, and immune compartments, including 45 immune subclusters with ncRNAs representing 24-38% of significantly differentially expressed genes.
Results:
To investigate the impact of ncRNAs driven by cytogenetic mutations, we first identified ncRNAs differentially expressed (P< 0.05) in myeloma cells between patients harboring specific mutations versus those without. Subsequent survival analyses revealed 29 cytogenetic-specific ncRNAs associated with patient outcomes (P< 0.01), including tumor suppressors (N=16, HR< 0.4) and oncogenes (N=13, HR >1.4). Network and pathway analyses indicated these ncRNAs may modulate critical cancer pathways, including proliferation and intercellular signaling. Differential abundance testing revealed that expression of 20 of these ncRNAs is associated with altered immune composition in 22 subpopulations (P< 0.05); for example, increased expression of amp(1q21)-associated oncogenic ncRNA HSALNG0008530 resulted in an increased proportion of mature neutrophils linked to poor survival. Lastly, to validate the clinical relevance of these ncRNAs, we are investigating their identities and functions in vitro.
Conclusions:
Collectively, our study presents a comprehensive coding-noncoding MM-BMME atlas, highlighting ncRNAs as regulators of MM phenotypes and immune microenvironment interactions, offering promising biomarkers and therapeutic targets.
