Myeloma Genomics and Microenvironment and immune profiling
Category: Myeloma Genomics and Microenvironment and immune profiling
Comparison of NGS-Based PlasmaSeq and FISH for Routine Genomic Testing in Multiple Myeloma
(PA-261) Comparison of NGS-Based PlasmaSeq and FISH for Routine Genomic Testing in Multiple Myeloma

Parth Shah, MD
Staff Physician
Dartmouth Hitchcock Medical Center
Fluorescence in situ hybridization (FISH) is widely used for cytogenetic risk stratification in multiple myeloma (MM) but has limitations due to restricted probe panels and high sample input requirements. Moreover, recent IMS/IMWG guidelines suggest the need to move over to sequencing based methodologies. PlasmaSeq is a next-generation sequencing (NGS)-based assay that detects copy number changes, structural variants, and relevant somatic mutations for risk-stratification and immunotherapy targets. This study compares FISH and PlasmaSeq in bone marrow (BM) samples from routine real world clinical testing.
Methods:
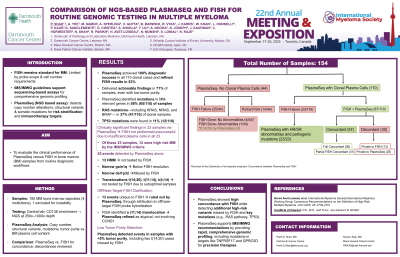
Fifty-nine BM samples from MM patients at various stages were analyzed by PlasmaSeq (NCGM, Raleigh NC) and standard FISH per institutional protocols. PlasmaSeq identified clonal plasma cell–associated genomic alterations including aneuploidy, deletions, amplifications, translocations, and mutations. Concordance was assessed in cases where both tests were performed; discordances were reviewed and categorized.
Results:
DNA input ranged from 1ng/ul to 90.6ng/ul with a bone marrow plasma cell content ranging from < 5% to ~80% across all samples. PlasmaSeq detected clonal plasma cells in 43/59 (73%) samples based on an integrated mutational profile based clonal determination tool. Of the remaining 16 where PlasmaSeq indicated no clonal profile, FISH failed in 14 and was negative in 2. Of the 43 patients, PlasmaSeq identified 18(42%) Hyperdiploid (HMM) cases. In 14 of 43 PlasmaSeq-positive cases (32.6%), FISH was either not performed or unsuccessful; all 14 had clinically significant findings on PlasmaSeq. Twenty-nine cases had results from both tests: 21 (72%) were concordant. Eight cases (28%) were discordant, with 7 abnormalities unique to PlasmaSeq while 2 were detected with FISH. However, the events specific to FISH were ruled out by sequencing (1p32 deletion mapped outside CDKN2C and gain1q located downstream of CKS1B outside of the chr1q21 cytoband). Of the 7 events detected by PlasmaSeq alone, 4 cases were HMM and not tested by FISH, a narrow gain1q event below FISH resolution, t(14;20) translocation and narrow del1p32 missed by FISH. Furthermore, PlasmaSeq was able to identify mutations in MM-relevant genes in 65% of patients. RAS pathway genes were involved in 18 patients(42%) and TP53 mutations in 6(14%) patients with clonal plasma cells.
Conclusions:
PlasmaSeq achieved a 100% diagnostic discovery rate in all 43 clonal-positive cases. In 32.6% (14/43), PlasmaSeq refined or reinterpreted FISH-reported abnormalities. These results support PlasmaSeq, a sequencing-based method, as an important tool for comprehensive genomic profiling in MM.
