Myeloma Genomics and Microenvironment and immune profiling
Category: Myeloma Genomics and Microenvironment and immune profiling
Ultra-deep, cost-efficient whole-genome sequencing of cell free DNA recovers most bone marrow mutations in newly diagnosed multiple myeloma
(PA-203) Ultra-deep, Cost-efficient Whole-genome Sequencing of Cell Free DNA Recovers Most Bone Marrow Mutations in Newly Diagnosed Multiple Myeloma

Dor D. Abelman, HBHSc, PhD(c) (he/him/his)
PhD Candidate
Princess Margaret Cancer Centre
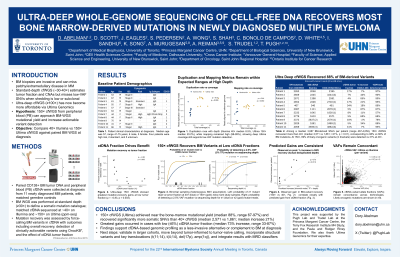
Cell-free DNA whole-genome sequencing (cfWGS) provides a non-invasive alternative to tumor biopsy for molecular profiling. Standard‑depth (~30–40×) cfWGS estimates tumour fraction and broad copy‑number alterations but often misses single‑nucleotide variants (SNVs) and small indels below 10% variant‑allele frequency. Recent studies in solid tumours have shown that pushing cfWGS to ultra‑deep coverage (≥100×) can rescue low‑frequency somatic mutations, yet this has not been explored in multiple myeloma (MM). With sequencing costs declining, ultra-deep cfWGS now holds promise for the near-complete genomic reconstruction of MM from peripheral blood (PB) cfDNA alone.
To determine whether ultra‑deep cfWGS can approach the mutational yield of matched bone-marrow (BM) WGS and capture clinically actionable variants, we compared standard‑depth (40×) Illumina cfWGS with 200× cfWGS generated on the new, low‑cost Ultima Genomics platform.
Methods:
Paired BM and PB cfDNA samples with matched buffy coat germline controls were obtained at diagnosis from eight transplant-eligible MM patients in the Multiple Myeloma Molecular Monitoring (M4) study. BM CD138+ DNA underwent 30-40× WGS on Illumina NovaSeq 6000. Matched cfDNA libraries were sequenced to 40× on Illumina (aligned with BWA-MEM/GATK with somatic variants called via MuTect2) and 200× on Ultima Solaris™ (prepared with PPM-Seq and processed through Ultima’s variant pipeline). cfDNA tumor fraction was estimated using ichorCNA (Adalsteinsson et al., 2017).
Results:
At diagnosis (median age 59, range 41-67; 5M/3F), 3/8 patients were high-risk, 2 standard-risk, and 3 unknown; subtypes included 4 IgG, 3 IgA, and 1 light-chain only. WGS of DNA derived from CD138+ selected plasma cells identified a median of 3,437 somatic mutations (range 633–4,681). In these untreated patients, cfDNA tumor fractions ranged from 4.5% to 33.7% (median 12.0%). Standard 40× cfWGS recovered a median of 1,654 mutations, while 200× cfWGS recovered 2,510 (range 455–4,153), representing a 48% median increase (range 13–102%). Relative to tumor-derived DNA, 200× cfWGS captured 80% of mutations and 69% of OncoKB actionable tier 1-2 variants (such as NRAS p.Q61R, KRAS p.A146V and TP53 p.M237I), compared to 56% and 46% at 40×, respectively. The percent increase in mutation recovery with ultra-deep sequencing correlated inversely with cfDNA tumor fraction (Spearman’s ρ = –0.88; p < 0.01). In all three samples with ≤5% tumor fraction, 200× cfWGS recovered at least 50% more tumor-identified mutations than 40×, with gains up to 102%.
Conclusions:
Ultra-deep cfWGS using Ultima sequencing recovered a median of 80% of tumor-derived mutations (range 65% to 89%) in this pilot cohort, supporting cfDNA as a standalone substrate for comprehensive MM genomic profiling and potentially reducing invasive BM sampling. Future work will validate these findings in larger cohorts and extend the analysis to other variant types including translocations.
